Antibody Libraries for In Vitro Display: What to Consider
The strength of any display-based antibody discovery program hinges on its library. While all library types have the potential to generate antibodies with high affinity and specificity, they differ in how reliably leads generated from the screening process can be converted into viable candidates during development. The diversity (quantity of and variety between library sequences), functionality (capability to produce viable antibodies), and developability (commercially relevant considerations including recombinant expression level, solubility, stability, and immunogenicity) of sequences in the library govern the likelihood of generating successful candidates. Antibody libraries are typically derived from one or a combination of natural or synthetic sources including naïve B-cell repertoires, immunized donors, and combinatorial DNA synthesis. To help you decide which type of library is the most appropriate for your in vitro display experiment, we’ll look at the preparation, benefits, and challenges of each one.
Naïve Antibody Libraries
Advantages:
- Functionally derived
- Human sourcing
- Faster production than immune (weeks)
Disadvantages:
- Library bias from donor selection
- Downstream optimization after screening
Compared to immunized repertoires, naïve libraries have a larger diversity as there is no or minimal antigen bias. The goal is to employ the full range of the human immunoglobulin repertoire for affinity maturation in vitro. These libraries are constructed from VH and VL sequences isolated from healthy human donors in a manner that is similar to immune libraries, minus the immunization step. They typically represent a pool of carefully selected donors to ensure a high diversity of sequences. Naïve libraries can be used to screen against a wide range of antigens; however, antibodies from these sources may be unpredictable for downstream developability. In any natural antibody library, toxicity to E. coli is not uncommon, making them incompatible with some popular screening technologies. Even candidates that exhibit favorable affinity and specificity through screening may also exhibit expression, solubility, or some other physiochemical or biological property that harms commercial viability. Consideration of the full scope of an antibody discovery and development program can avoid costly and time-intensive optimization downstream.
Immune Antibody Libraries
Advantages:
- Functionally derived
- Well-established and proven method
Disadvantages:
- Slow to produce (months)
- Non-human sourcing
- Low compatibility with contemporary screening/development
Antibody repertoires derived from natural sources have the advantage of practical functionality that has been demonstrated in vivo. They can be isolated from immunized or non-immunized (naïve) sources with repertories of human or non-human origin. Using immune libraries for a display approach enables antibody selection both in vivo and in vitro. The adaptive immune system enriches B-cells that produce specific and functional antibodies against the antigen, and this population of immunoglobulin (Ig) sequences is further optimized through directed evolution via in vitro display. Library generation typically involves isolating B-cells from an immunized individual and cloning the variable regions of the heavy and light antibody chains into a vector (Figure 1). When originating from a non-human repertoire, antibody drugs carry an elevated risk of eliciting undesired immune responses in patients despite “humanization” of the antibody sequences post-selection. However, using transgenic animals—in which endogenous Ig genes have been replaced with human counterparts—significantly mitigates this risk. The genetic bias inherent in immune libraries effectively restricts the range of sequences that are screened in vitro, which has benefits and drawbacks. The reduced diversity improves the chances of isolating a functional candidate but limits the potential gains in affinity from in vitro selection.
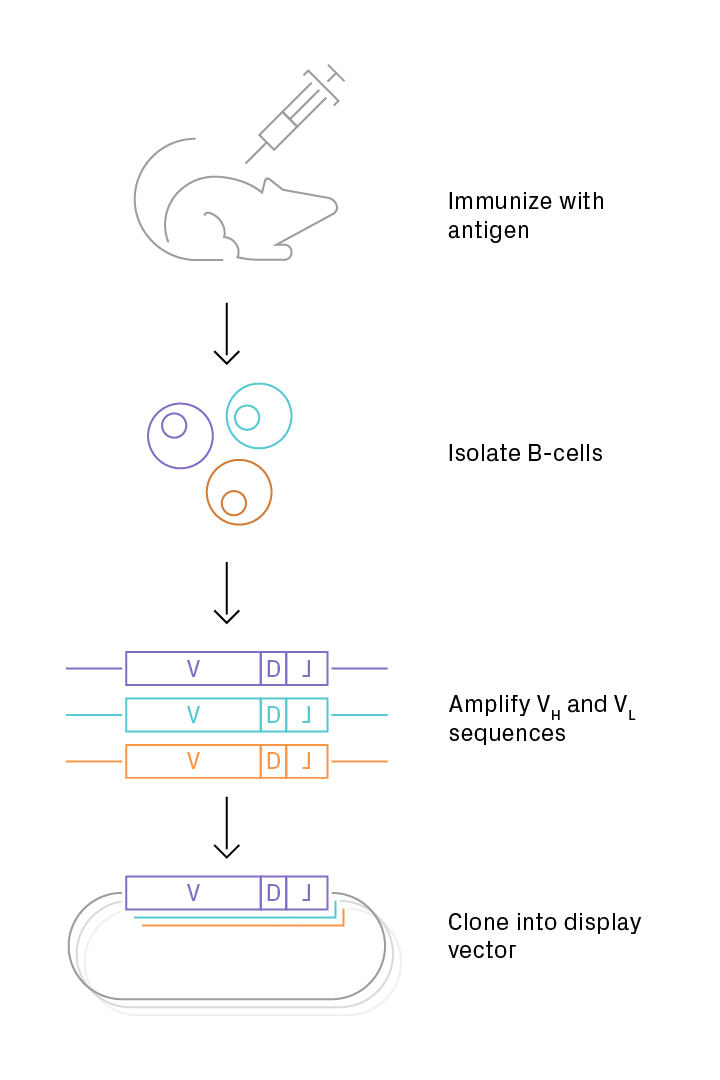
Figure 1. Preparation of an immune antibody library. The target protein, or antigen, is introduced into an animal. Later, B-cells are subsequently isolated, and mRNA is purified. The V(D)J sequences are amplified (via RT-PCR) from the immunoglobulin heavy- and light-chain variable regions and cloned into a vector, such as a phagemid for phage display.
Synthetic Antibody Libraries
Advantages:
- High degree of library control
- Optimized meaningful diversity
- Faster production than immune libraries (weeks)
- Rational design based on project goals
Disadvantages:
- Additional laboratory work required for synthesis
For the purposes of this discussion, we will focus exclusively on antibody libraries generated through chemical synthesis/modification rather than semi-synthetic repertoires. These libraries are designed in silico using data from a variety of sources including natural repertoire sequences and then constructed using current DNA synthesis technologies. Offering a high degree of control, this approach enables a “quality by design” approach that favors development. In a synthetic repertoire, molecular properties that are critical to commercial success can be a focal point of the library by using pre-optimized scaffolds from literature. Diversification takes place in targeted positions to maximize a range of binder selectivity while minimizing disruption to folding or other structural elements. This means that naturally variable domains such as the complementary determining regions (CDRs) are selectively mutated using a method such as combinatorial DNA synthesis, while framework regions are conserved to minimize developability biases.
| Combinatorial Library | Stop Codons | Risk of Mutation | ‘AT’ Rich Bias | Assigned Amino Acids | Screening Speed | Screening Costs |
|---|---|---|---|---|---|---|
| Random (NNN) | 3 | High | Possible | No | Slow | $$$ |
| Partially Random (NNK/NNS) | 1 | High | Possible | No | Moderate | $$ |
| Trimer-Controlled | 0 | Low | None | Yes | Fastest | $ |
Table 1. Comparison of combinatorial synthetic DNA library techniques
The most common and simple method of CDR diversification uses randomized (NNN) or degenerate (NNK/NNS) mixtures of codons to create variants of a parent DNA sequence coding for an antibody (Table 1). Unfortunately, these methods can introduce stop codons or frameshift mutations into the library, creating a nonviable antibody and increasing the time and costs required for subsequent screening. Additionally, the randomized nature of oligonucleotide use in NNN/NNK/NNS libraries results in low precision for sequence diversification. Trinucleotide-directed mutagenesis (TRIM) or other trimer-controlled DNA synthesis techniques use pre-synthesized codon units to allow precise control of amino acid distribution and limitation of nonviable antibodies due to stop codon incorporation or disruptions to antibody folding from frameshift mutations. A smart synthetic method such as trimer-control enables precise construction of antibody libraries through deliberate protein design and codon optimization. Synthetic errors such as insertions or deletions can be minimized by using high-quality oligonucleotide building blocks and optimized DNA assembly protocols. A diversity analysis and quality control check of the library by Sanger or next-generation sequencing provides confidence in the final product.
Key Takeaways
- Several factors should be considered when building an antibody library, including sequence diversity and the predictability of developing antibody leads (from in vitro screening) into viable candidates
- Immune and naïve library generation techniques are proven to be effective for specific antigens, but possess inherent limitations for developability
- Antibody libraries produced from synthetic DNA can be quickly and easily engineered to fit the biological and commercial goals of a development program
See how a trimer-controlled synthesis approach can boost the effectiveness of antibody libraries by downloading our case study.
References
1. Shim, H. Synthetic approach to the generation of antibody diversity. BMB Rep. 2015, 48, 489-494.
2. Almagro, J. C.; Pedraza-Escalona, M.; Arrieta, H. I.; Pérez-Tapia, S. M. Phage Display Libraries for Antibody Therapeutic Discovery and Development. Antibodies 2019, 8, 44-65.
3. Lin, C.-W.; Lerner, R.A. Antibody Libraries as Tools to Discover Functional Antibodies and Receptor Pleiotropism. Int. J. Mol. Sci. 2021, 22, 4123-4134.
4. Chan S.K., Rahumatullah A., Lai J.Y., Lim T.S. (2017) Naïve Human Antibody Libraries for Infectious Diseases. In: Lim T. (eds) Recombinant Antibodies for Infectious Diseases. Advances in Experimental Medicine and Biology, vol 1053. Springer, Cham. https://doi.org/10.1007/978-3-319-72077-7_3
Daily Insight
Recent Posts
Recent Post Tags
- next generation sequencing
- gene synthesis
- rna sequencing
- sanger sequencing
- transcriptomics
- aav-itr
- itr sequencing
- proteomics
- single-cell sequencing
- aav plasmid prep
- antibody discovery
- cell and gene therapy
- olink
- quantitative pcr (qPCR)
- rna therapeutics
- PacBio
- circular rna (circRNA)
- digital pcr (dPCR)
- lentivirus
- long-read sequencing
- mRNA
- oxford nanopore sequencing (ont)
- pcr + sanger sequencing
- whole plasmid sequencing